Archives
- 2025-12
- 2025-11
- 2025-10
- 2023-07
- 2023-06
- 2023-05
- 2023-04
- 2023-03
- 2023-02
- 2023-01
- 2022-12
- 2022-11
- 2022-10
- 2022-09
- 2022-08
- 2022-07
- 2022-06
- 2022-05
- 2022-04
- 2022-03
- 2022-02
- 2022-01
- 2021-12
- 2021-11
- 2021-10
- 2021-09
- 2021-08
- 2021-07
- 2021-06
- 2021-05
- 2021-04
- 2021-03
- 2021-02
- 2021-01
- 2020-12
- 2020-11
- 2020-10
- 2020-09
- 2020-08
- 2020-07
- 2020-06
- 2020-05
- 2020-04
- 2020-03
- 2020-02
- 2020-01
- 2019-12
- 2019-11
- 2019-10
- 2019-09
- 2019-08
- 2019-07
- 2019-06
- 2019-05
- 2019-04
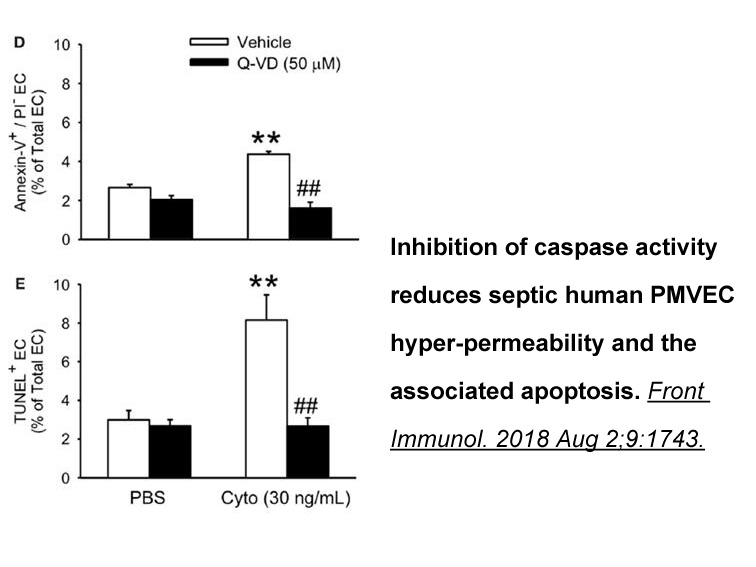
- 2018-07
-
br Interpretation of identified genetic
2019-05-27
Interpretation of identified genetic variants and genetic counseling Although the current expert consensus statement recommends that comprehensive or BrS1 (SCN5A) targeted BrS genetic testing can be useful for genetic testing [63], not all the published SCN5A mutations in BrS have been subjected
-
En el cuento el protagonista
2019-05-27
En el cuento, el protagonista es sometido propyl los abusos de la mercantilización del objeto artístico y de una distorsionada crítica de arte que buscan en él diversas explicaciones o justificaciones a su propia razón de ser a través de mecanismos legitimadores, enmarcados en el entorno posvanguard
-
El n mero de correspondiente al primer semestre de ofrece
2019-05-27
El número 58 de correspondiente al primer semestre de 2014, ofrece la sección Movimientos sociales y democracia en América Latina, en la cual se encuentran temas de gran relevancia política y social para nuestra América. En primer lugar, el artículo “Sentidos de lugar y movimiento social: indígenas
-
Resulta entonces anacr nico por parte
2019-05-27
Resulta entonces anacrónico, por parte de Loustaunau, culpar fda approved Ca rrión de la migración masiva de ecuatorianos a España a finales del siglo y decir que tal migración “es la rebelión de un pueblo dispuesto a actuar por sí mismo, a pesar de las consecuencias, y así dejar de ser manipulado
-
The deletion of q seen in
2019-05-27

The deletion of 14q seen in our patient included the IGH locus at 14q32.33. This is noteworthy, because loss of 14q has been reported as a recurrent aberration acquired during the natural history of CLL. The acquisition of a del(14) may thus be indicative of CLL in transformation. In a study of a la
-
br Discussion Our results confirm that
2019-05-27
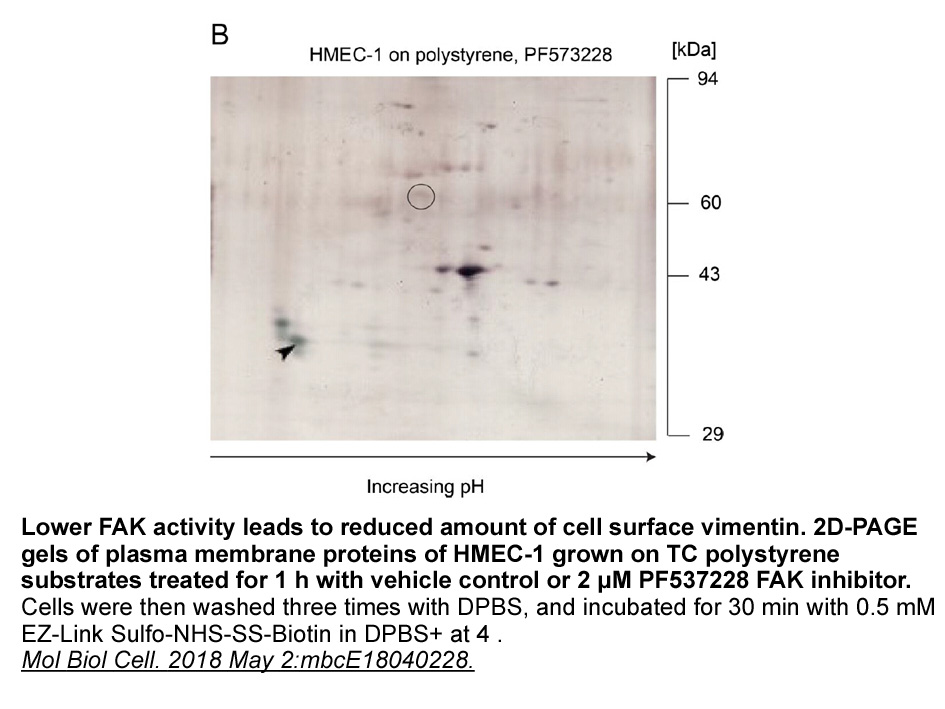
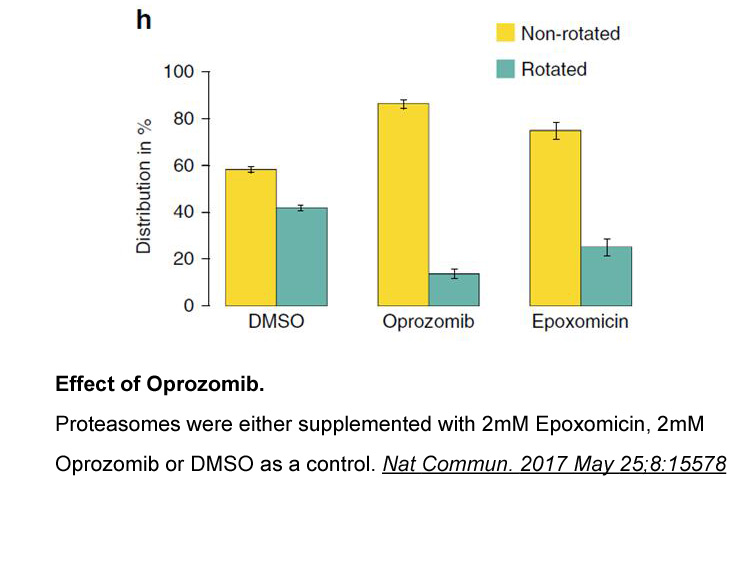
Discussion Our results confirm that germline variations in the TPMT gene are associated with treatment outcome in children with ALL [5]. The ability of TPMT to catalyze S-methylation and inactivate 6-mercaptopurine makes it a major determinant of 6-mercaptopurine toxicity. Furthermore, reduced TP
-
A total of microorganism species in
2019-05-27
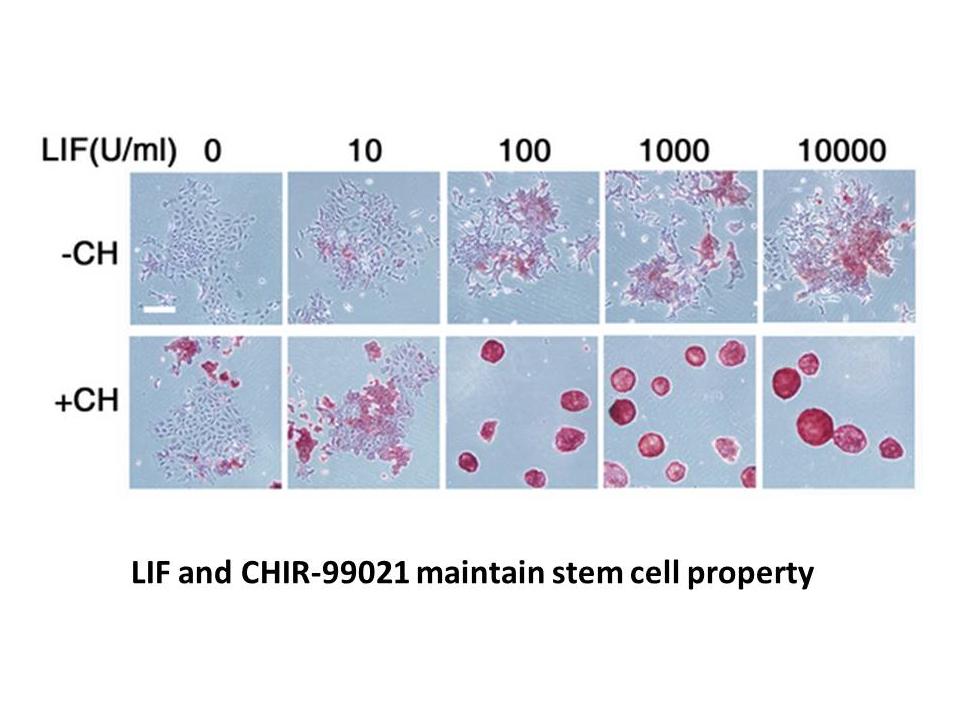
A total of 46 microorganism species in the old NICU group and 35 in the new NICU group were isolated, respectively. Klebsiella pneumoniae was the most frequent pathogen in the old NICU group (Table 4), followed by E. coli, Methicillin-resistant Staphylococcus aureus (MRSA), Coagulase-negative Staphy
-
Almost all men are eligible to have
2019-05-24
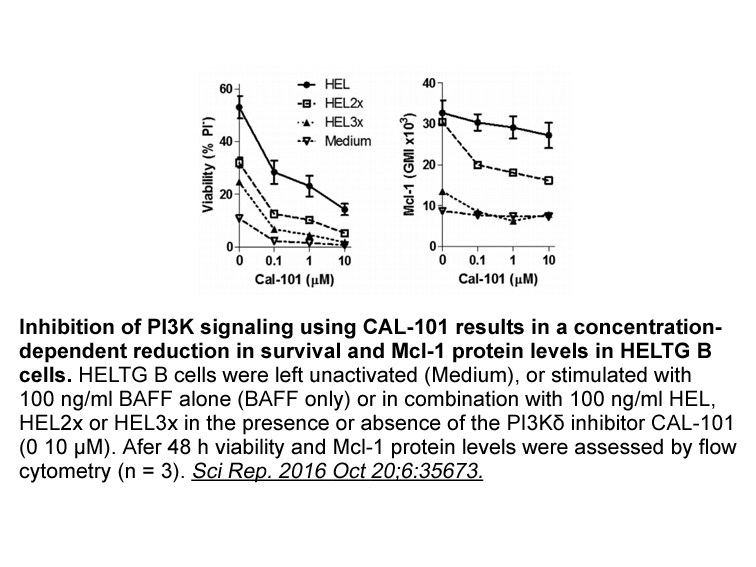
Almost all men are eligible to have a vasectomy. It is highly effective, convenient, and easy to provide, and is also, along with female sterilisation, one of only two permanent methods of contraception. For these reasons, and to share contraceptive responsibilities, anti fungal is widely chosen in
-
pak 4 Fischer and colleagues also raise concerns about the
2019-05-24
Fischer and colleagues also raise concerns about the fact that MHSP “only offers testing to men”. As previously stated, MHSP is a public health programme based on the principles outlined in WHO\'s interim guidelines. The focus of that guidance is preventing sexual transmission of Ebola virus by men
-
Introduction Cancer therapy has evolved throughout the decad
2019-05-24
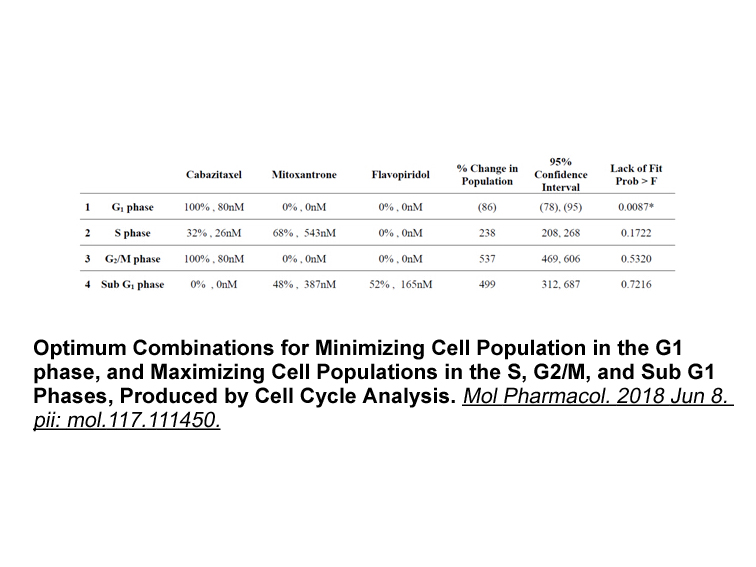
Introduction Cancer therapy has evolved throughout the decades and is beginning to branch off into new and exciting fields including immunotherapy. The hope of this new field stems from the lack of success of chemotherapy in general. Chemotherapy\'s cytotoxic characteristics were regarded as one of
-
In the American College of Chest Physicians
2019-05-24

In 2013, the American College of Chest Physicians provided a guideline to the preoperative physiological assessment of patients being considered for surgical resection of lung cancer. It has been recommended that patients must be assessed by a multidisciplinary team before operation, regardless of a
-
It has been speculated that
2019-05-24
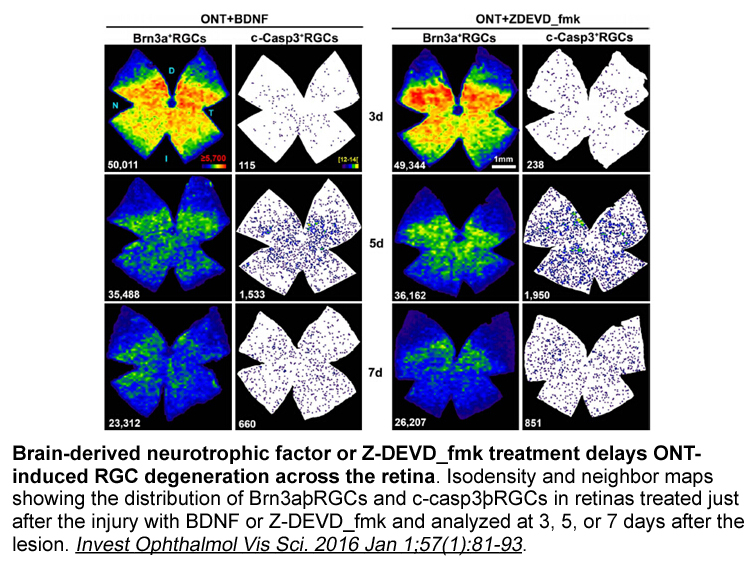
It has been speculated that the geographical variation among radiotherapy prescription is a function of the respective healthcare systems within each region. If physicians receive greater reimbursement for multiple fractions compared to a single fraction, they will likely have greater willingness to
-
Recent studies have analyzed the miRNA profile of serum
2019-05-24
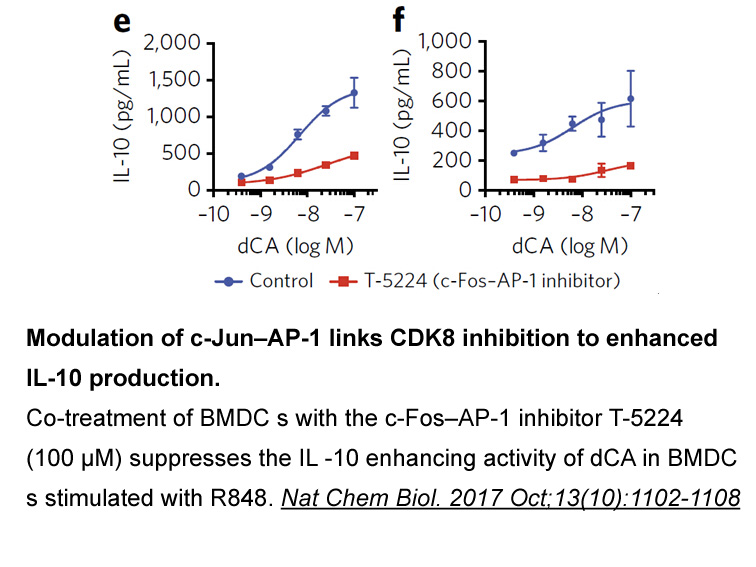
Recent studies have analyzed the miRNA profile of serum and bile samples from patients with PSC and patients with CCA complicating PSC. Overall, the miRNA expression of all analyzed miRNAs from both patients with PSC and patients with CCA complicating PSC was significantly higher compared with healt
-
br Drug resistant tuberculosis threatens to undermine advanc
2019-05-24
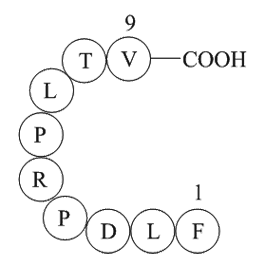
Drug-resistant tuberculosis threatens to undermine advances in tuberculosis control worldwide. Multidrug-resistant tuberculosis (MDRTB), defined as resistance to isoniazid and rifampicin, requires treatment for up to 24 months with expensive diagnostics and second-line drugs. Fewer than 15% of pat
-
br A gest o hospitalar nas unidades mantidas Talvez
2019-05-24
A gestão hospitalar nas unidades mantidas Talvez seja o produto de mais expressividade que as oss divulgam. A forma como se gerencia o hospital é abacavir sulfate garantia de colheita dos resultados, seu game over junto à sociedade e junto a sua contratante, a Secretaria de Saúde. É a procura por
11241 records 731/750 page Previous Next First page 上5页 731732733734735 下5页 Last page